Covid 19 - État de l'art
L’émergence d'un nouveau coronavirus, SARS-CoV-2, en Chine, en décembre 2019, est à l’origine d’une épidémie mondiale, source d’inquiétudes et de questionnements, souvent justifiés mais parfois aussi démesurés. En cette période de confinement, nous sommes inondés d'informations et de prises de paroles, qui peuvent se révéler anxiogènes, inexactes ou encore volontairement polémiques. Cependant, de nombreuses études scientifiques sont publiées quotidiennement et souvent en accès libre : elles peuvent nous permettre de faire la part des choses et de mieux saisir les enjeux de cette épidémie de COVID-19.
EspritsCo vous propose un modeste état de l'art des travaux et études menés par la communauté scientifique mondiale avec un premier volet consacré aux principales questions concernant SARS-CoV-2 et la maladie qu'il induit COVID-19.
1. SARS-CoV-2 : caractéristiques morphogénétiques et origines
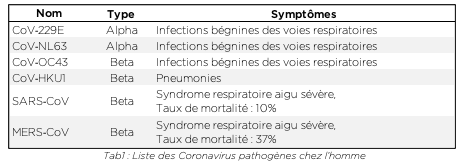
Les Coronavirus (CoV) sont une famille de virus possédants un génome à ARN très long. Ils sont entourés d’une enveloppe sphérique recouverte de protéines qui leur donnent une forme caractéristique de couronne, à laquelle ils doivent leur nom. Il existe de nombreux sous-types de coronavirus infectant différentes espèces animales. Au moins six d’entre eux sont capables d’infecter l’homme, les plus courants étant HCoV-229E et HCoV-OC43 (voir tab.1). Le plus souvent, ces virus sont associés à des rhumes et des symptômes grippaux bénins. Chez un certain nombre de sujet, ils n’induisent même aucuns symptômes. Cependant, ils peuvent également être responsables de complications respiratoires sévères (pneumopathies), telles que des pneumonies, en particulier chez des sujets dits sensibles[1] (maladies chroniques, personnes âgées…).
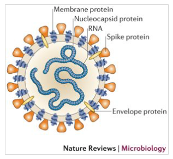
Ce n’est pas la première fois qu’une épidémie de coronavirus touche l’espèce humaine. Le SARS-CoV (SRAS = syndrome respiratoire aigu sévère) et le MERS-CoV ont déjà provoqué deux épisodes épidémiques mortels à l’échelle mondiale respectivement en 2003 et en 2012. Nous utiliserons régulièrement ces deux coronavirus comme référence dans le cadre de cet article.
SARS-CoV-2 appartient à cette grande famille de virus, c’est un béta-coronavirus[2] constitué d'une enveloppe protéique de 60 à 220nm de diamètre. Cette enveloppe renferme un ARN linéaire simple brin d’environ 29.000 nucléotides qui a été rapidement séquencé en tout début d’année 2020. La séquence génétique de ce Coronavirus est similaire à 82% à celle du SARS-CoV[3] d’où son nom SARS-CoV-2.
D’un point de vue phylogénétique, le SARS-CoV-2 est très proche des séquences virales de coronavirus présentes chez la chauve-souris[4]. Ces chiroptères, qui représentent près de 20% de la diversité des mammifères, sont connus pour constituer un réservoir important de virus[5,6,2] et leur distribution mondiale en font un candidat parfait pour les zoonoses (maladies humaines d’origine animale). Cependant, pour que cette transmission inter-espèce puisse avoir lieu, il est indispensable qu’un certain nombre de mutations génétiques interviennent afin de permettre au coronavirus de remplir deux conditions essentielles :
- Être reconnu par les récepteurs présents à la surface des cellules humaines pour les infecter
- Être capable de se transmettre d’homme à homme.
SARS-CoV constitue un modèle particulièrement pertinent pour comprendre les paramètres[2] qui ont pu permettre à SARS-CoV-2 de franchir les espèces pour infecter l’homme et engendrer la pandémie que nous connaissons actuellement.
SARS-CoV est issu de la même région que SARS-CoV-2 : le sud de la Chine et plus précisément le marché aux animaux de la province de Guangzhou. Ce n’est pas un hasard si deux pandémies de Coronavirus ont eu pour foyer cette région de la Chine : on y trouve un certain nombre de marchés aux animaux qui, combinés à une densité humaine importante et des conditions de captivité peu hygiéniques, constituent l’incubateur parfait pour l’émergence de nouveaux virus[7,8]. C’est ainsi que, dans la province chinoise de Guangzhou, SARS-CoV est notamment passé de la chauve-souris à la civette palmiste masquée de l’Himalaya puis à l’homme selon un phénomène stochastique aléatoire[9,10]. Autrement dit, le marché aux animaux de Guangzhou a très probablement été le lieu de multiples contaminations inter-espèces lors desquelles, les mutations indispensables à une transmission humaine ont été naturellement sélectionnées[11–13,9,14].
Cependant, si l’environnement est une condition nécessaire à l’émergence de ce type de virus, elle n’est pas suffisante : c’est également la plasticité génétique des coronavirus qui a permis l’émergence de SARS-CoV en 2003 et de SARS-CoV-2 à la fin 2019. Les coronavirus montrent un taux de mutation, associé à la réplication de leur ARN simple brin, particulièrement important au regard des autres rétrovirus. Cette plasticité offerte par leur complexe de réplication (notamment RdRp) est particulièrement efficace du fait de la longue taille de leur ARN qui permet de limiter les pertes fonctionnelles liées aux mutations. Enfin, cette plasticité est encore amplifiée par la faculté du virus à recombiner son ARN au contact d’autres virus dans le cadre d’une poly-infection[2]. Il a été démontré que dans le cadre de SARS-CoV, le virus était parvenu à s’adapter à de nombreux hôtes en particulier via des mutations au niveau du domaine RBD (receptor binding domain) de sa protéine S qui ont notamment permis la fixation du virus au récepteur ACE2 des cellules de la sphère ORL d’un certain nombre de mammifères, dont l’homme[14,15].
Ce sont fort probablement ces mécaniques qui ont permis l’émergence de SARS-CoV-2 au marché de Wuhan. La communauté scientifique suspecte le pangolin[16–18] d’avoir joué le rôle d’hôte intermédiaire bien que cette hypothèse reste à confirmer. La maladie causée par le SARS-CoV-2 depuis décembre 2019 a été nommée de manière définitive Covid-19 (Corona virus disease) le 11 février 2020 par l’OMS.
2. COVID-19 : diagnostic, modes de transmission, symptômes et taux de mortalité
2.1. Détection et localisation du virus chez les sujets infectés
Comment fonctionnent les tests de dépistage COVID-19 ?
La mesure de la charge virale par RT-PCR est l’approche la plus fiable pour déterminer la localisation du virus et par conséquent la manière dont nous nous le transmettons. C’est également principalement sur cette méthode d’analyse que reposent les tests qui permettent de déterminer si un individu a été infecté par le virus par le biais d’un prélèvement nasopharyngé[19,20].
Les différents prélèvements réalisés par les équipes médicales depuis le début de l’épidémie semblent indiquer pour le moment que le virus se situe :
- Principalement dans le système respiratoire[21]
- Dans les selles jusqu’à 27 jours après les 1ers symptômes[22]
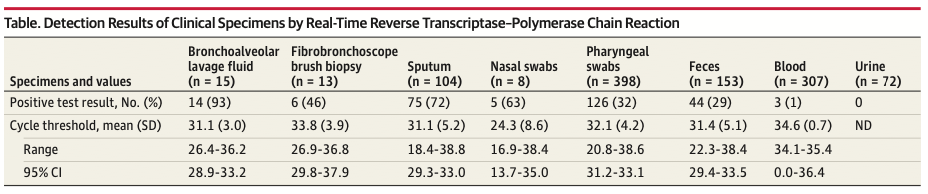
Tab 2 : Detection of SARS-CoV-2 in Different Types of Clinical Specimens, JAMA, march 11, 2020 [23]
Dans une étude menée à l’hôpital universitaire de Nanchang[24] (Chine), du 21 janvier au 4 février 2020 sur 76 patients atteints de COVID-19, les auteurs ont monté que la charge virale moyenne des cas graves était en moyenne 60 fois supérieure à celle des cas bénins. Les cas bénins se sont révélés avoir une clairance virale précoce, avec 90% de ces patients testés négatifs 10 jours après l’infection. En revanche, tous les cas graves sont restés positifs au jour 10 et les jours qui ont suivi, avec une détection du virus dans le sang des patients les plus graves[25].
Les tests sont-ils fiables ? En existe-t-il d’autres ?
Les résultats présentés en Tab.2 montrent une détection de SARS-CoV-2 par RT-PCR dans 63% des cas chez des patients COVID-19 suite à un prélèvement nasopharyngé. Ils posent par conséquent la question de la sensibilité des tests de dépistage : une étude en particulier a montré que ces tests peuvent se monter négatifs chez des patients présentant un tableau clinique typique[26]. Le recours à une analyse des selles par RT-PCR a finalement permis de détecter la présence du virus.
Si la RT-PCR permet de détecter la présence de certains gènes caractéristiques de SARS-CoV-2 chez les patients testés, d’autres tests de type ELISA [27] peuvent également permettre de dépister les patients. Ils reposent sur la détection des protéines virales et peuvent être complétés par une analyse de type Western Blot pour éliminer tout doute sur le statut du patient (ce type d’approche est notamment réalisée dans le cadre du dépistage du VIH).
Nous verrons plus tard dans le second volet de ce dossier que ces tests de dépistage occupent une place centrale tant dans les stratégies mises en place par certains pays pour lutter contre l’épidémie que, fort probablement, dans les stratégies de déconfinement qui interviendront post pic épidémique.
2.2. Une contagiosité relativement importante
Comment évalue-t-on la contagiosité ?
Le calcul de la contagiosité de COVID-19 est fondé sur le taux de reproduction de base, appelé R0. Ce taux de reproduction représente le nombre moyen de nouvelles infections induites par une personne porteuse de SARS-CoV-2.
A titre d’exemple, un R0 de 4 signifie que chaque sujet infecté va contaminer à son tour en moyenne 4 nouvelles personnes. Ainsi, lorsque le R0 est supérieur à 1, la maladie va s’étendre dans la population en absence d’actions sanitaires alors que lorsque le R0 est inférieur à 1, la maladie va s’éteindre d’elle-même.
A titre indicatif, voici quelques valeurs de R0 associés à des maladies connues :
- Grippe saisonnière : 1,3[28]
- SARS-CoV : entre 2 et 4[29]
- MERS-CoV : entre 2 et 5[30]
L'OMS estime que le R0 du SARS-CoV-2 est compris entre 1,4 et 2,5, quand la majorité des publications scientifiques affiche une valeur moyenne de 2,7[29,31].
D’autres facteurs peuvent également être intéressants à prendre en compte dans le cadre de l’estimation de la contagiosité, notamment :
- Le délai de contagiosité pré-symptomatique[32] : 24-48h
- La période d’incubation[33,34] : 1-14 jours avec une médiane à 5-6j[35]
La combinaison d’une période de contagiosité pré-symptomatique de l’ordre d’1 ou 2 jours[36], d’un R0 relativement élevé tout au long de la période d’incubation et de formes peu symptomatiques voire asymptomatiques de la maladie est probablement responsable de la cinétique rapide d’évolution de l’épidémie à l’échelle mondiale[37,38].
Existe-t-il des super-contaminateurs ?
Difficile d’aborder la question de la contagiosité de COVID-19 sans mentionner le concept de « super contaminateur » qui a alimenté un certain nombre d’articles de presse[1,2] et de discussions sur les médias sociaux. "Ce n'est pas un terme médical […] il a été utilisé pour désigner une personne qui infecte un nombre proportionnellement très important d'individus, sans qu'il y ait forcément un seuil précis », déclarait récemment à l’AFP Amesh Adalja, médecin spécialiste des maladies infectieuses émergentes et de la préparation aux pandémies, à l'Université américaine Johns Hopkins.
Ce concept n’est pas nouveau et avait déjà suscité un certain nombre de questions lors des précédentes épidémies de SARS et de MERS sans que sa véracité n’ait vraiment fait l’objet de travaux de recherche. Deux individus : l’homme d’affaire britannique Steve Walsh et la sexagénaire coréenne surnommée patiente 31, ont récemment relancé les débats dans le cadre de COVID-19.
Le professeur Eric Caumes, chef du service des maladies infectieuses et tropicales à l'hôpital parisien de la Pitié-Salpêtrière et le Pr Olivier Bouchaud, chef du service des maladies infectieuses de l'hôpital Avicenne de Bobigny se sont également exprimés sur le sujet : s’ils ne réfutent pas l’hypothèse de l’existence d’individus capables d’en contaminer de nombreux autres, ils indiquent qu’il n’existe pas de preuve ni d’explication formelles sur cette question. Le Dr Bharat Pankhania, spécialiste des maladies infectieuses de la faculté de médecine de l'Université britannique d'Exeter, préfère parler de situation de super propagation (« super spreading event ») qui est beaucoup plus consensuel dans la communauté scientifique[39] et qui correspond à une conjonction de facteurs propices à une contamination massive (ventilation, rassemblement d’individus dont un au moins est au début de la période de contagiosité…).
2.3. Modes de transmission
Au-delà de l’évident risque de contamination liés aux contacts de proximité avec des individus infectés (inhalation des gouttelettes, contact avec les sécrétions d’un individu infecté), deux autres questions animent les débats autour des modes de transmission :
- La question de la stabilité du virus sur des surfaces inertes : une fois déposé sur un matériau donné, combien de temps le virus est-il capable d’infecter un nouvel individu ?
- La question des risques d’infections liés à des contacts indirects avec ces surfaces inertes : quelle quantité de virus minimale doit être présente sur ces surfaces pour qu’un contact indirect main-muqueuse (nasale, oculaire ou buccale) entraine une infection ?
La communauté scientifique mondiale tente, entre autres, de répondre à ces questions cruciales afin de mieux comprendre la dynamique de propagation de l’épidémie. Une étude relativement récente a tenté d’estimer le temps durant lequel le virus restait détectable sur des surfaces inertes[40] :
- Plastique : de 7 à 10h pouvant aller jusqu'à 3 jours
- Acier inoxydable : de 6 à 9h pouvant aller jusqu'à 3 jours
- Carton : de 4 à 6 heures pouvant aller jusqu'à 1 jour
- Cuivre : de 1 à 2h pouvant aller jusqu'à 4 heures
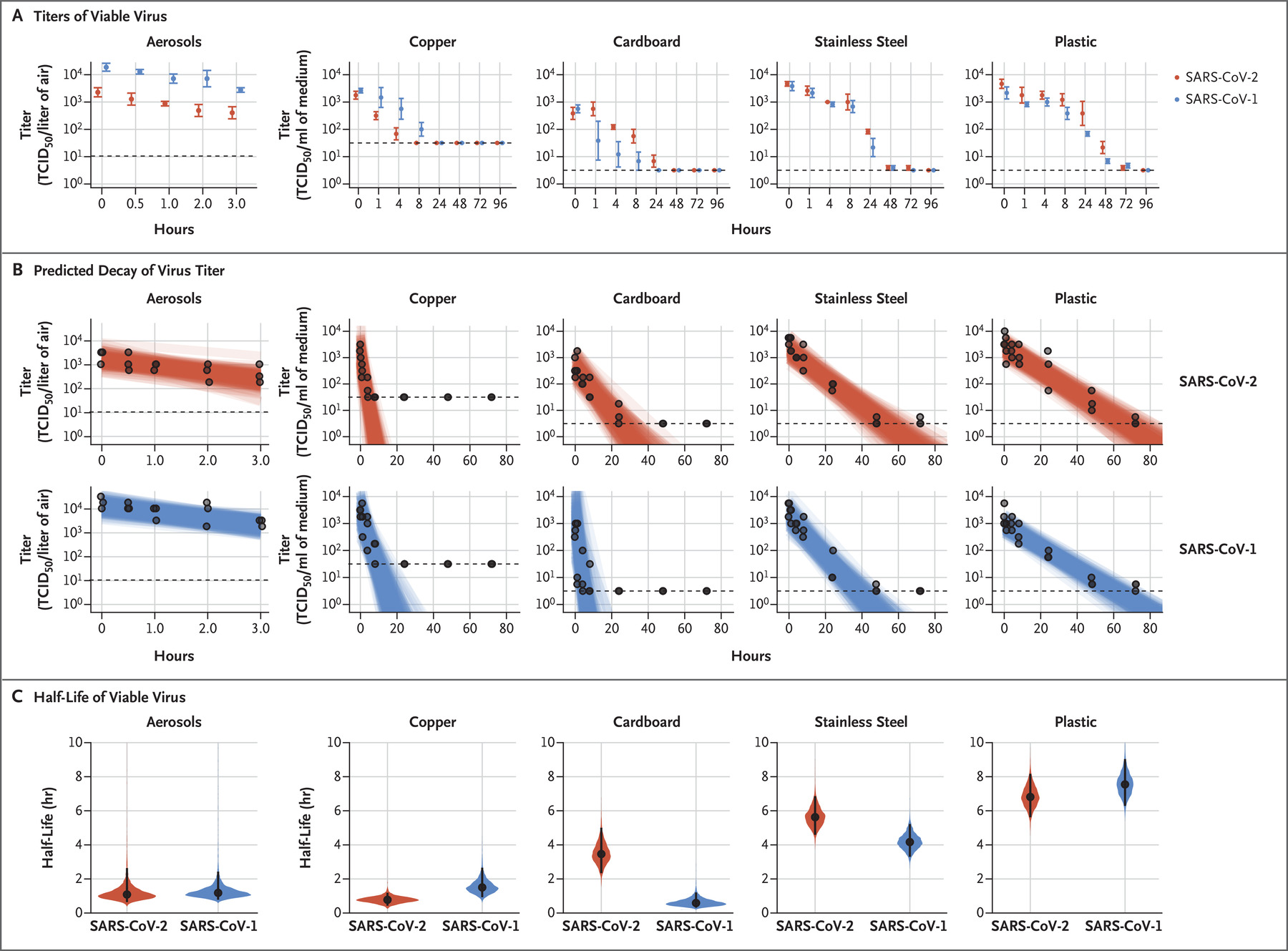
En revanche, concernant le risque de contagion que le contact avec ces surfaces souillées représente, la virologue Astrid Vabre chef du service de virologie du CHU de Caen et spécialiste des coronavirus, indiquait dans une interview menée par un journaliste du quotidien le Monde que « Ce qui pose vraiment problème, c’est qu’on ne connaît pas la dose infectante ». Si le manque de données ne permet pas de déterminer avec prévision la dose de virus nécessaire pour contaminer un individu, les différences interindividuelles pourraient également probablement jouer un rôle (état des muqueuses, système immunitaire, fatigue…).
2.4. Des symptômes qui s’apparentent à ceux d’une grippe saisonnière
Quels sont les symptômes de COVID-19 ?
Les symptômes de la maladie s’apparentent pour la plupart à des symptômes grippaux[41]. Cependant, les cas les plus sévères aboutissent à des pneumopathies ainsi que des défaillances rénales. Voici les principaux symptômes répertoriés ainsi que leur fréquence d’apparition chez les individus infectés[42] :
- Une fièvre modérée chez 87 % des sujets infectés.
- Une toux sèche dans 60% des cas
- Une myalgie (fatigue) pour 39% des sujets contaminés
- Une dyspnée (troubles respiratoires) dans 18% des cas
- Des douleurs musculaires et articulaires dans 15% des cas
- Des maux de gorge et des céphalées (maux de tête) dans 14% des cas souvent accompagnés d’une congestion et d’écoulement nasal
- Des manifestations gastro-intestinales, en particulier des diarrhées[43]
De récentes études ont également mis en avant d’autres symptômes tels que :
- Une anosmie[44,45] et une dysgueusie[46] (perte du goût et de l’odorat)[47,48] pour 86% des patients concernés
- Des manifestations dermatologiques[49] ou oculaires[50].
Quel est le tableau clinique des cas graves de COVID-19 ?
Le tableau clinique des patients hospitalisés et présentant des formes graves de la maladie[42] est le suivant :
- Taux élevé de protéine C réactive chez 99% des malades
- Lymphopénie (réduction du nombre de leucocytes) chez 60% des patients.
- Au scanner : détection d’une atteinte pulmonaire chez 90% des patients admis à l’hôpital, avec des lésions communes de forme inégales, une opacité du verre dépolie au scanner chez 60% des patients, un épaississement vasculaire dans 80% des cas, un signe de halo pour 64% d’entre eux.
Une étude[25] publiée dans the Lancet Infectious Diseases et réalisée par les équipes du service des maladies infectieuses et tropicales des hôpitaux Bichat (Paris) et du CHU de Bordeaux sur 5 patients suggère l’existence de 3 typologies de patients COVID-19 :
- Les patients présentant une forme bégnine de la maladie : pauci symptomatiques, ils se caractérisent par une évolution spontanée rapidement favorable malgré une forte charge virale mesurée après prélèvement nasophanryngé. Ces résultats semblent confirmer les résultats d’une étude réalisée en Chine[21] qui soulignaient la contagiosité de ce type de patients.
- Les patients biphasiques : Ces patients présentent des symptômes modérés durant les 10 premiers jours de la maladie suivis d’une aggravation subite sur le plan respiratoire malgré une baisse de la charge virale dans leur prélèvements nasopharyngés. Les auteurs de l’étude indiquent que les lésions pulmonaires observées chez ces patients pourraient être induits par une réponse immunitaire excessive (phénomène pro-inflammatoire très important) plutôt que par la progression du virus.
- Les patients graves : les symptômes sont d’emblée importants avec une évolution rapide de la maladie induisant des défaillances multi- viscérales (détresse respiratoire, insuffisance rénale…). Ces patients présentent une charge virale élevée tout au long de la maladie, non seulement dans les prélèvements nasopharyngés mais également dans les selles et dans le sang notamment. Le plus souvent cette typologie de patients concerne des individus âgés et présentant des facteurs de comorbidité.
Les statistiques mondiales indiquent que dans 80% les cas, les symptômes de COVID-19 sont modérés, légers voire inexistants, ils sont en revanche sévères dans 15% des cas et critiques dans 5% des cas. Ainsi, 20% des individus infectés par COVID-19 nécessitent une hospitalisation et l’on estime qu’en moyenne 3% des patients infectés meurent du COVID-19 dans les services de réanimation[51]. Cependant, la mortalité du COVID-19 pose encore question et ne pourra véritablement être déterminée qu’en testant massivement les populations afin de prendre en compte, notamment, les individus pauci-symptomatiques qui jouent fort probablement un rôle marqué dans la propagation du virus.
2.5. Une mortalité surestimée et hétérogène dans le monde
Nous savons aujourd’hui que le Covid-19 est le plus souvent bénin en l'absence de pathologie chronique préexistante[52] (hypertension, diabète, maladies cardiovasculaire, cancers, etc.). L’étude la plus large réalisée à ce jour, par le Centre de Contrôle Des maladies Chinois (CCDC - Chinese Center for Disease Control and Prevention), portant sur 45000 sujets atteints de COVID-19, montre que le risque de décès augmente avec l’âge[51] :
- Aucun décès n’a à ce jour été constaté chez les enfants de moins de 10 ans,
- Le risque est de l’ordre de 0,2% chez les sujets d’entre 10 et 39 ans,
- Il passe à 0,4% chez les quadragénaires,
- Il s’élève à 1,3% chez les quinquagénaires,
- Il monte à 3,6% chez les sexagénaires,
- Il atteint 8% chez les septuagénaires
- Il est le plus fort chez les octogénaires et atteint les 14,8%
Les résultats de cette étude ont été mis en perspective avec ceux publiés en Italie[53] :
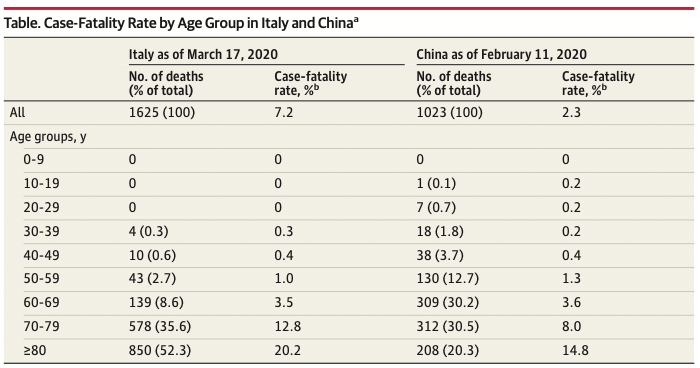
Ces résultats convergent en ce qui concerne « l’âge charnière » de 60 ans à partir duquel, la mortalité augmente significativement, elle est particulièrement marquée chez les 70-90 ans.
Ces résultats divergent quant aux taux de mortalité globaux observés dans les 2 pays. Ils permettent de souligner 2 points importants :
- Les taux de mortalité de COVID-19 définis quotidiennement sont probablement peu significatifs en l’absence de dépistage systématique de la population[54].
- La mortalité est hétérogène dans le monde[55].
Si la question des biais d’estimation de la mortalité fait consensus, plusieurs arguments ont été avancés pour expliquer son hétérogénéité :
- L’hétérogénéité de la distribution des cas dans les pays[53] corrélée à l’importance des décès chez les patients de plus de 80 ans. La démographie des pays pourrait expliquer en partie les différences observées.
- La surcharge induite par l’épidémie dans les services hospitaliers, en particulier dans les services de réanimation[55]
- Les stratégies de dépistages qui diffèrent considérablement d’un pays à l’autre : l’Asie du sud Est a testé massivement ses populations alors que les pays Européens ont eu tendance à rationaliser l’utilisation des tests (du fait de la tension importante sur les consommables notamment).
Pour aller plus loin
1. Les sites d’information de référence
- SRLF - Société de réanimation de langue Française
- INSERM - Dossier d’information Coronavirus et COVID-19
- Société Française de pharmacologie et de thérapeutique
- Santé Publique France
- Infovac-France : la plateforme d’information sur la vaccination
- Direction Générale de la Santé,
- OMS – Organisation Mondiale de la santé
- CDC – Center for disease control and prevention
- Institut John Hopkins
2. Suivre l’évolution de la pandémie dans le monde
- Infographie de la cinétique de propagation de COVID-19 dans le monde et en France par santé publique France.
- Suivi quotidien de l’évolution de l’épidémie dans le monde à partir des chiffres consolidés de l’université Johns Hopkins.
- Covid-19 Dashboard par Pratap Vardhan fondé sur les chiffres de Johns Hopkins.
3. Vulgarisation scientifique
- chronique de Nicolas Martin sur France culture "Radiographies du corona virus"
- Collectif de bénévoles (majoritairement issus de la recherche en biologie) travaillant à la production de contenus vulgarisés sur la #Covid19 et le #SarsCov2
Références bibliographiques
1. Chen Y, Liu Q, Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418-423. doi:10.1002/jmv.25681
2. Chan JF-W, To KK-W, Tse H, Jin D-Y, Yuen K-Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21(10):544-555. doi:10.1016/j.tim.2013.05.005
3. Chan JF-W, Kok K-H, Zhu Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221-236. doi:10.1080/22221751.2020.1719902
4. Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270-273. doi:10.1038/s41586-020-2012-7
5. Fan Y, Zhao K, Shi Z-L, Zhou P. Bat Coronaviruses in China. Viruses. 2019;11(3):210. doi:10.3390/v11030210
6. Hu B, Zeng L-P, Yang X-L, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. Drosten C, ed. PLOS Pathog. 2017;13(11):e1006698. doi:10.1371/journal.ppat.1006698
7. Guan Y. Isolation and Characterization of Viruses Related to the SARS Coronavirus from Animals in Southern China. Science. 2003;302(5643):276-278. doi:10.1126/science.1087139
8. Lau SKP, Woo PCY, Li KSM, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci. 2005;102(39):14040-14045. doi:10.1073/pnas.0506735102
9. Song H-D, Tu C-C, Zhang G-W, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci. 2005;102(7):2430-2435. doi:10.1073/pnas.0409608102
10. Graham RL, Donaldson EF, Baric RS. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013;11(12):836-848. doi:10.1038/nrmicro3143
11. Tu C, Crameri G, Kong X, et al. Antibodies to SARS Coronavirus in Civets. Emerg Infect Dis. 2004;10(12):2244-2248. doi:10.3201/eid1012.040520
12. The Chinese SARS Molecular Epidemiology Consortium. Molecular Evolution of the SARS Coronavirus During the Course of the SARS Epidemic in China. Science. 2004;303(5664):1666-1669. doi:10.1126/science.1092002
13. Yang Z -y., Werner HC, Kong W -p., et al. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc Natl Acad Sci. 2005;102(3):797-801. doi:10.1073/pnas.0409065102
14. Bolles M, Donaldson E, Baric R. SARS-CoV and emergent coronaviruses: viral determinants of interspecies transmission. Curr Opin Virol. 2011;1(6):624-634. doi:10.1016/j.coviro.2011.10.012
15. Graham RL, Baric RS. Recombination, Reservoirs, and the Modular Spike: Mechanisms of Coronavirus Cross-Species Transmission. J Virol. 2010;84(7):3134-3146. doi:10.1128/JVI.01394-09
16. Zhang T, Wu Q, Zhang Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr Biol. 2020;30(7):1346-1351.e2. doi:10.1016/j.cub.2020.03.022
17. Wong MC, Javornik Cregeen SJ, Ajami NJ, Petrosino JF. Evidence of Recombination in Coronaviruses Implicating Pangolin Origins of NCoV-2019. Microbiology; 2020. doi:10.1101/2020.02.07.939207
18. Xiao K, Zhai J, Feng Y, et al. Isolation and Characterization of 2019-NCoV-like Coronavirus from Malayan Pangolins. Microbiology; 2020. doi:10.1101/2020.02.17.951335
19. Chu DKW, Pan Y, Cheng SMS, et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin Chem. 2020;66(4):549-555. doi:10.1093/clinchem/hvaa029
20. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3). doi:10.2807/1560-7917.ES.2020.25.3.2000045
21. Zou L, Ruan F, Huang M, et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020;382(12):1177-1179. doi:10.1056/NEJMc2001737
22. Wu Y, Guo C, Tang L, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5(5):434-435. doi:10.1016/S2468-1253(20)30083-2
23. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. March 2020. doi:10.1001/jama.2020.3786
24. Liu Y, Yan L-M, Wan L, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. March 2020:S1473309920302322. doi:10.1016/S1473-3099(20)30232-2
25. Lescure F-X, Bouadma L, Nguyen D, et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. March 2020:S1473309920302000. doi:10.1016/S1473-3099(20)30200-0
26. Chen L, Lou J, Bai Y, Wang M. COVID-19 Disease With Positive Fecal and Negative Pharyngeal and Sputum Viral Tests: Am J Gastroenterol. March 2020:1. doi:10.14309/ajg.0000000000000610
27. Guo L, Ren L, Yang S, et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin Infect Dis. March 2020:ciaa310. doi:10.1093/cid/ciaa310
28. Biggerstaff M, Cauchemez S, Reed C, Gambhir M, Finelli L. Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: a systematic review of the literature. BMC Infect Dis. 2014;14(1):480. doi:10.1186/1471-2334-14-480
29. Liu Y, Gayle AA, Wilder-Smith A, Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020;27(2):taaa021. doi:10.1093/jtm/taaa021
30. Choi S, Jung E, Choi BY, Hur YJ, Ki M. High reproduction number of Middle East respiratory syndrome coronavirus in nosocomial outbreaks: mathematical modelling in Saudi Arabia and South Korea. J Hosp Infect. 2018;99(2):162-168. doi:10.1016/j.jhin.2017.09.017
31. Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. The Lancet. 2020;395(10225):689-697. doi:10.1016/S0140-6736(20)30260-9
32. Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med. 2020;382(10):970-971. doi:10.1056/NEJMc2001468
33. Ganyani T, Kremer C, Chen D, et al. Estimating the Generation Interval for COVID-19 Based on Symptom Onset Data. Infectious Diseases (except HIV/AIDS); 2020. doi:10.1101/2020.03.05.20031815
34. Nishiura H, Linton NM, Akhmetzhanov AR. Serial interval of novel coronavirus (COVID-19) infections. Int J Infect Dis. 2020;93:284-286. doi:10.1016/j.ijid.2020.02.060
35. Li Q, Guan X, Wu P, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N Engl J Med. 2020;382(13):1199-1207. doi:10.1056/NEJMoa2001316
36. Yu P, Zhu J, Zhang Z, Han Y. A Familial Cluster of Infection Associated With the 2019 Novel Coronavirus Indicating Possible Person-to-Person Transmission During the Incubation Period. J Infect Dis. February 2020:jiaa077. doi:10.1093/infdis/jiaa077
37. Bai Y, Yao L, Wei T, et al. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA. 2020;323(14):1406. doi:10.1001/jama.2020.2565
38. Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science. March 2020:eabb3221. doi:10.1126/science.abb3221
39. Han Y, Yang H. The transmission and diagnosis of 2019 novel coronavirus infection disease (COVID‐19): A Chinese perspective. J Med Virol. March 2020:jmv.25749. doi:10.1002/jmv.25749
40. van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N Engl J Med. March 2020:NEJMc2004973. doi:10.1056/NEJMc2004973
41. Wang D, Hu B, Hu C, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061. doi:10.1001/jama.2020.1585
42. Han R, Huang L, Jiang H, Dong J, Peng H, Zhang D. Early Clinical and CT Manifestations of Coronavirus Disease 2019 (COVID-19) Pneumonia. Am J Roentgenol. March 2020:1-6. doi:10.2214/AJR.20.22961
43. Gu J, Han B, Wang J. COVID-19: Gastrointestinal Manifestations and Potential Fecal–Oral Transmission. Gastroenterology. March 2020:S001650852030281X. doi:10.1053/j.gastro.2020.02.054
44. Brann DH, Tsukahara T, Weinreb C, et al. Non-Neuronal Expression of SARS-CoV-2 Entry Genes in the Olfactory System Suggests Mechanisms Underlying COVID-19-Associated Anosmia. Neuroscience; 2020. doi:10.1101/2020.03.25.009084
45. Iacobucci G. Sixty seconds on . . . anosmia. BMJ. March 2020:m1202. doi:10.1136/bmj.m1202
46. Eliezer M, Hautefort C, Hamel A-L, et al. Sudden and Complete Olfactory Loss Function as a Possible Symptom of COVID-19. JAMA Otolaryngol Neck Surg. April 2020. doi:10.1001/jamaoto.2020.0832
47. Gautier J-F, Ravussin Y. A New Symptom of COVID-19: Loss of Taste and Smell. Obesity. April 2020. doi:10.1002/oby.22809
48. Vaira LA, Salzano G, Deiana G, De Riu G. Anosmia and Ageusia: Common Findings in COVID-19 Patients: Otolaryngological Manifestations in COVID-19. The Laryngoscope. April 2020. doi:10.1002/lary.28692
49. Recalcati S. Cutaneous manifestations in COVID-19: a first perspective. J Eur Acad Dermatol Venereol. March 2020. doi:10.1111/jdv.16387
50. Wu P, Duan F, Luo C, et al. Characteristics of Ocular Findings of Patients With Coronavirus Disease 2019 (COVID-19) in Hubei Province, China. JAMA Ophthalmol. March 2020. doi:10.1001/jamaophthalmol.2020.1291
51. Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239. doi:10.1001/jama.2020.2648
52. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet. 2020;395(10229):1054-1062. doi:10.1016/S0140-6736(20)30566-3
53. Onder G, Rezza G, Brusaferro S. Case-Fatality Rate and Characteristics of Patients Dying in Relation to COVID-19 in Italy. JAMA. March 2020. doi:10.1001/jama.2020.4683
54. Baud D, Qi X, Nielsen-Saines K, Musso D, Pomar L, Favre G. Real estimates of mortality following COVID-19 infection. Lancet Infect Dis. March 2020:S147330992030195X. doi:10.1016/S1473-3099(20)30195-X
55. Ji Y, Ma Z, Peppelenbosch MP, Pan Q. Potential association between COVID-19 mortality and health-care resource availability. Lancet Glob Health. 2020;8(4):e480. doi:10.1016/S2214-109X(20)30068-1
